Why do we sleep?
Why we sleep remains one of the most enduring mysteries in science. Sleep is homeostatically regulated where the duration of wakefulness drives subsequent sleep. Here we aim to determine how waking experience is sensed to trigger sleep and how sleep restores the brain? How the circadian clock interacts with sleep homeostasis? How impaired sleep leads to human diseases such as Alzheimer’s disease?
why do we sleep?
Why we sleep remains one of the most enduring mysteries in science. Sleep is homeostatically regulated where the duration of wakefulness drives subsequent sleep. Here we aim to determine how waking experience is sensed to trigger sleep and how sleep restores the brain? How the circadian clock interacts with sleep homeostasis? How impaired sleep lead to human diseases such as Alzheimer’s disease?
Research
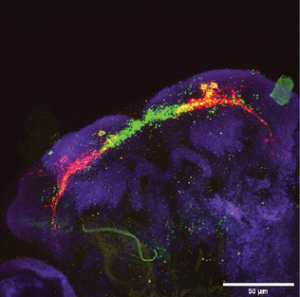
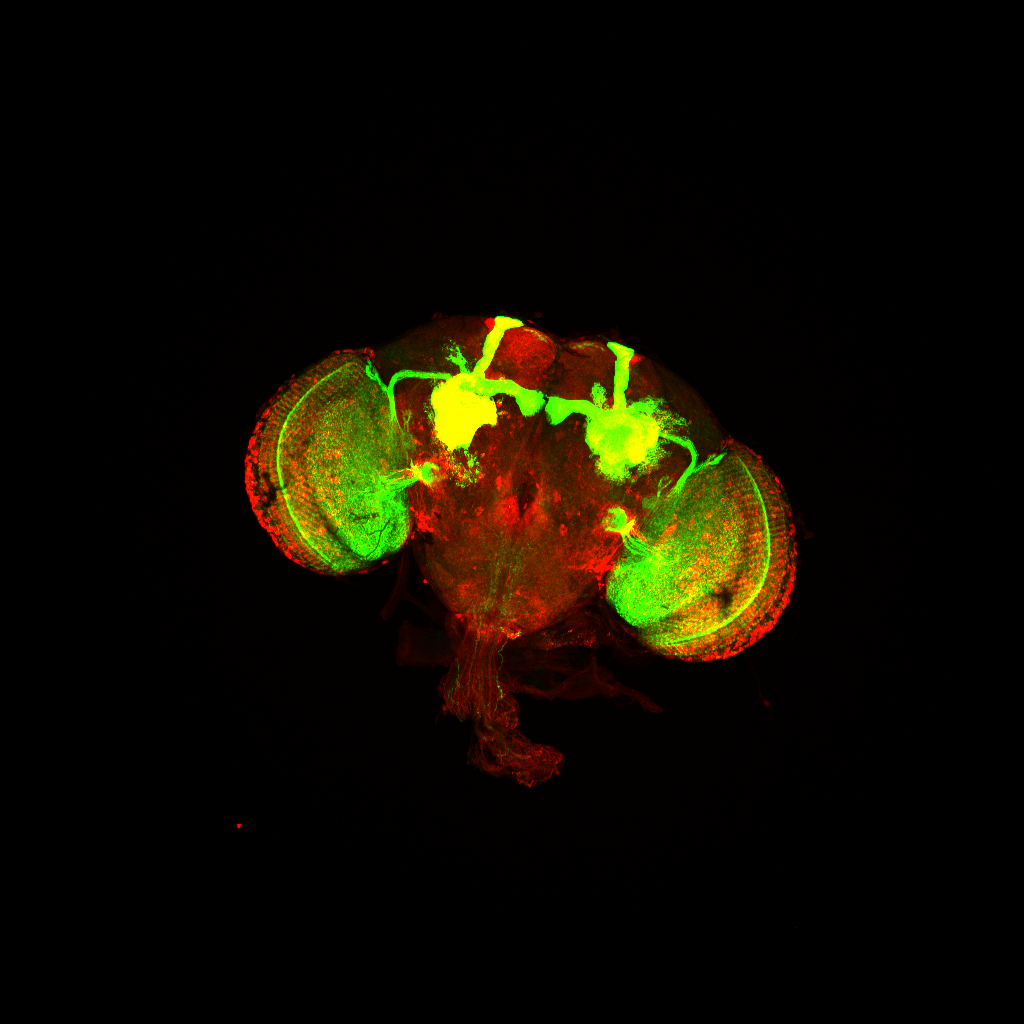
Nearly every organism examined, even the jellyfish that lacks a centralized nervous system, exhibits a restorative sleep-like state. While asleep, we cannot eat, mate, defend ourselves from predators or care for our young. Inadequate sleep contributes to brain disease such as Alzheimer’s and depression, and even diseases outside of the brain, such as diabetes and obesity. Sleep is homeostatically regulated, i.e., sleep is driven by the duration and intensity of prior waking experience. Our principal goals are to identify the neuronal and molecular components of the sleep homeostat, to understand how those components cooperate to sense and control sleep-wake state, and to reveal how molecular and neural homeostatic pathways impact brain function, health, and disease. Our strategy employs genetic tools primarily in the fruit fly Drosophila to dissect, manipulate and monitor the homeostatic machinery. This approach builds on our research to understand the molecular basis of circadian (~24 h) behavior which have revealed sleep mechanisms conserved between invertebrates and vertebrates and incorporates work in both mice and humans.
Circadian Rhythms
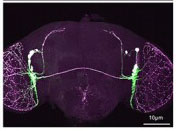
Circadian clocks dictate when we wake up and when we fall asleep. Using molecular to behavioral approaches primarily in the fruit fly we aim to reveal the molecular and neuronal mechanisms by which circadian clocks keep time and convey that information to control sleep/wake? We are also determining the mechanistic role of disrupted circadian clocks in neurodevelopmental (e.g. autism) and neurodegenerative (e.g. Huntington’s disease disorders.
Sleep Homeostasis
The elusive sleep homeostat drives sleep as a function of prior wakefulness. How does the homeostat sense waking experience, trigger sleep and restore the brain to a baseline healthy state? By combining genetics, genomics, real-time imaging, and high resolution behavior analysis, we aim to identify the locus of the sleep homeostat and understand the molecular mechanisms that govern homeostat function. We are also determining how disrupted sleep can contribute to neurodegenerative disorders such as Alzheimer’s disease.

Humans: Diagnostics/Jet Lag
Disrupted sleep and circadian rhythms have been increasingly associated with neural (depression and Alzheimer’s) and even non-neural disorders (diabetes and obesity). Here we are combining RNA-sequencing with machine learning algorithms to discover biomarker signatures of sleep and circadian disorders. We are also using publicly available data to assess the effects of sleep/circadian disruption on athletic performance.
Team

Ravi Allada
Principal Investigator
BA, University of Michigan, Ann Arbor
MD, University of Michigan, Ann Arbor
On his way to becoming a medical doctor, Ravi decided to take a year (and then two) off during medical school to do research at the NIH. There he caught the research ”bug” and began his career-long interest in sleep, working on mechanisms of general anesthesia in the fruit fly Drosophila. After completing his M.D and a short residency in Clinical Pathology, he did his postdoctoral training with Nobel laureate Michael Rosbash, cloning the core circadian clock gene Clock in Drosophila. He joined the faculty at Northwestern in 2000 where his lab has discovered core gears of the circadian clock, how those gears drive sleep and wake, and how those pathways are linked to neurodegenerative disease. He applies similar approaches to reveal the molecular basis of the sleep homeostat, key to understanding the elusive function of sleep. In his spare time, he has served or is serving on various Boards including for the Society for Research on Biological Rhythms, the Sleep Research Society and the NIH Sleep Disorders Research Advisory Board.

Rajesh Narasimamurthy
Assistant Research Scientist
MSc, Bharathidasan University, India
PhD, University of Zurich, Switzerland
Rajesh completed his PhD at the University of Zurich, Switzerland, where he studied signaling mechanisms of the Drosophila Tumour Necrosis Factor (TNF), Eiger. In his postdoctoral research at the Salk Institute, La Jolla, USA, he discovered a novel molecular interaction between the circadian clock and inflammation system. He then worked as a Group Leader at the Thermo Fisher Scientific, Bangalore, India, in the development and production of recombinant antibodies. Later, Rajesh returned to academic research at the Duke-NUS Medical School, Singapore, where he investigated the dynamic role of CK1 (Casein Kinase 1) and PER2 (Period 2) proteins in regulating molecular mechanisms of the circadian clock. Currently, he is interested in investigating the mechanisms of sleep homeostasis and circadian clock and its implications in neuro degenerative diseases using proteomic and phosphoproteomic approaches in fly and mammalian models.

Farheen Akhtar
Research Fellow
BS, MS, Aligarh Muslim University, India
Ph.D., Aligarh Muslim University, India
Farheen earned her Ph.D. in Biotechnology from Aligarh Muslim University, India (2022), where she developed nanoparticle-based photodynamic therapies for multidrug-resistant infections, resulting in an Indian patent. She then pursued postdoctoral research at the University of Pennsylvania, investigating circadian rhythms and metabolic regulation in Drosophila and mouse models. After three years at Penn, she has joined the Allada Lab at the Michigan Neuroscience Institute, University of Michigan, where her work focuses on sleep homeostasis and its metabolic underpinnings. She is passionate about bridging basic biology with translational therapeutics to address complex biomedical challenges through interdisciplinary science.

Shiju Sisobhan
Research Area Specialist
PhD, Indraprastha Institute of Information Technology, Delhi
BTech, Cochin University of Science and Technology, Kochi
I grew up in the beautiful state of Kerala (India) where I received my bachelor’s and master’s degree in electronics engineering. Then I relocate to Delhi, where I did my Ph.D. in computational biology under Dr. K. Sriram. My work in Ph.D. studies focused on the mathematical modeling of the mammalian circadian rhythm. Then I worked as postdoc at Northwesern University, Chicago and at University of Michigan. In the Allada lab, I am focusing on developing computational tools for analyzing sleep and circadian behavior of Drosophila and human being. I am also working on bioinformatics analysis on bulk and single cell RNA sequencing data, machine learning and biomarker discovery.

Gregory Wesseling
Research Specialist, Lab Manager
MS, Grand Valley State University, Allendale
BS, Central Michigan University, Mount Pleasant
In graduate school at GVSU, Greg dissected larval drosophila brains to investigate histamine expression in deletion mutants generated through transposon-excision mutagenesis. When Greg joined the Allada lab, he dissected adult drosophila brains to investigate PER expression. Greg now works on project ENCODE to help document where transcription factors act in the fly genome. Before graduate school, Greg spent a summer at the University of Cambridge to study evolution. In his spare time, Greg enjoys watching college football and basketball.

Aldeb Perera
Ph.D. Candidate
BS, University of Illinois at Chicago
A Belizean native, Aldeb moved to Chicago in 2013 where he completed is BS in Biochemistry at the University of Illinois at Chicago. After graduating college, Aldeb worked in Dr. Wei Qiu’s lab at Loyola University Chicago for two years investigating liver cancer, after which he began his PhD work at Northwestern University in the Fall of 2020. He joined the Allada lab the following year, where he currently uses molecular biology and omics approaches to deconvolute sleep homeostasis. Outside of lab, Aldeb serves as the president of Northwestern’s SACNAS chapter, studies classical piano at Northwestern’s Bienen School of Music, and is a member of NEIU’s Judo Club. In his free time, he enjoys videogames, time outdoors, and game nights with his siblings.

Julia Kravchenko
Ph.D. Candidate
BS, University of Michigan, Ann Arbor
Julia completed her BS in Neuroscience in 2022 at UMich, where she discovered her love of research as an honor thesis student in Dr. Kent Berridge’s lab. After graduating, she worked as a research technician in Dr. Joanna Mattis’s lab studying the role of neural circuitry in seizure severity. In 2023, Julia matriculated to NGP, where under the mentorship of Drs Catherine Kaczorowski and Ravi Allada she now studies the genetic basis of sleep disruptions in Alzheimer’s disease using flies, mice, and humans. She’s happy to talk about fiber art, fantasy novels, and powerlifting.

Tuhin Chakraborty
Research Specialist
Ph.D., National Center for Biological Sciences (NCBS), Bangalore, India
MS, Viswa-Bharati University, Santiniketan, India
Dr. Tuhin Subhra Chakraborty is a Research Specialist at the Michigan Neuroscience Institute (MNI). He received a master’s degree in Zoology from Visva-Bharati University, Santiniketan, India and the Ph.D. in Neurobiology from the National Center for Biological Sciences (NCBS), Bangalore, India. After completing a postdoctoral fellowship in System Neuroscience at the University of California, San Diego, Tuhin joined the Department of Molecular and Integrative Physiology (MIP), University of Michigan in Ann Arbor where he uses Drosophila melanogaster as a model organism to study the impacts of severe experiences on an organism’s survival and quality of life. This is achieved through unraveling links between perception and homeostatic control.

Madison Hoogstra
Research Associate
BS, Calvin University, Grand Rapids
Maddy was born and raised near Chicago, where her passion for science first took root. She earned her B.S. in Biochemistry with a concentration in Neuroscience from Calvin University, gaining research experience through internships at both Calvin and the Van Andel Institute. After graduating, she joined the Allada Lab, where she applies molecular biology approaches to study sleep homeostasis in Drosophila. Outside the lab, Maddy can be found hiking, baking, rock climbing, or spending time with friends and family.

Aadish Mithil Shah
Associate Data Scientist
BSE Data Science, University of Michigan, Ann Arbor (c/o 2021)
Aadish was born and raised in Bahrain but is of Indian descent. After moving to the US in 2017, he completed my BSE in data science eng. at UMich, unofficially focusing in AI, ML and computational methods. After graduating in 2021, he spent three years building a social networking startup, launching 8 software products, fueled by pre-seed funding secured through the Desai Accelerator program- an entrepreneurship investment arm of the Zell Lurie Institute at the Ross School of Business. At the Allada Lab, Aadish leverages his data science and entrepreneurial experience to develop data applications that analyze fly scRNA/bulk RNA seq transcriptomics, model fly behavior, and automate experiments. His fascination with sleep and its profound impact on biology, inspired by personal experience, has deepened through his work at the Allada Lab, where he is driven to better understand the science behind why we sleep. Aside from work, Aadish also enjoys preparing meals for family & friends, playing the guitar and watching soccer games (especially when Chelsea is winning.)
Alumni
Postdoctoral Fellows
Clark Rosensweig Ph.D.
Sumit Saurabh Ph.D.
Dae-Sung Hwangbo, Ph.D.
Mikhail Koksharov Ph.D.
Evrim Yildirim Ph.D.
Jongbin Lee Ph.D.
Matthieu Flourakis Ph.D.
Taichi Itoh Ph.D.
Elzbieta Kula-Eversole Ph.D.
Tae Hee Han Ph.D.
Yong-Jae Kwon, Ph.D.
Chunghun Lim Ph.D.
Cory Pfeiffenberger Ph.D.
Gang Liu Ph.D.
Bridget Lear Ph.D.
Jena Pitman-Leung, Ph.D.
Elaine Smith Ph.D.
J. Russel Keath, Ph.D.
Jui-Ming Lin Ph.D.
Valerie Kilman Ph.D.
2017-2024
2018-2020
2012-2019
2016-2018
2014-2018
2012-2018
2009-2017
2012-2015
2008-2015
2012-2014
2012-2014
2007-2013
2007-2012
2007-2010
2002-2009
2002-2007
2002-2009
2003-2008
2001-2007
1998-2000
Operations Director, Single Cell Genomics Core, Northwestern University, IL
Assistant Professor, Chicago State University, IL
Assistant Professor, Biology, University of Louisville, KY
Senior Scientist, Synlife, Cambridge, MA
Postdoctoral Fellow, Neurology, University of Michigan, MI
Research Faculty, National Institute of Science & Technology, South Korea
Team Lead, Strategic Partnership & Stakeholder Management, Akkodis, Belgium
Assistant Professor, Experimental Natural Science, Kyushu University, Japan
Research Associate, Michigan State University, Grand Rapids, MI
Staff Scientist, National Institutes of Health (NIH), Washington DC
Senior Research Specialist, University of Illinois, IL
Professor, National Institute of Science & Technology, South Korea
Senior Scientific Manager, Global Medical Communications at ICON, PA
Corporate Vice President & Actuary, New York Life Insurance Company, NY
Research Associate Professor, Northwestern University, Highland Park, IL
Assistant Director, Neuroscience Program, Northwestern University, IL
Scientific Director, AbbVie, Chicago, IL
Scientist, Charles River Laboratories, Cleveland OH
R&D Staff Sccientist, IDEXX Laboratories, ME
Associate Professor, Assistant Director of Neuroscience, Northwestern University, IL
Ph.D. Students
Melanie Zhang, Ph.D.
Tomas Andreani, Ph.D.
Fangke Xu, Ph.D.
Adam Seluzicki, Ph.D.
Brian Chung, Ph.D.
Luoying Zhang, Ph.D.
Rose-Anne Meissner, Ph.D.
Kevin Keegan, Ph.D.
2015-2021
2014-2022
2012-2019
2007-2013
2006-2010
2004-2009
2004-2009
2003-2005
Medical Student, Medical Scientist Training Program, Feinberg School of Medicine, IL
Scientist II, Sarepta Therapeutics, MA
Researcher, Center for Brain Science, RIKEN, Japan
Research Associate, Chory Lab, Salk Institute, CA
Director of Programmatic Grants Development, Medical College of Wisconsin, WI
Professor, Chair of Science & Technology Department of Life Science, Huazhong University, Wuhan
Biomedical Editor, General Dynamics Information Technology (GDIT), TX
Independent Scientist, Big Data Analyst and Science Writer, IL
Master’s Students
Austin Wen, M.S.
Zhichun Lin, M.S.
Jack Qin, M.S.
Wenhao Cao, M.S.
Timothy Earl, LL.D.
Ling Kai Shih, M.S.
Zachary Taub, M.D.
Anuja Ganesan, M.S.
Weichao Zhang, Ph.D.
Matthew Hong, M.S.
Samuel Stewart, M.S.
Keya Raychaudhuri, M.D.
Sihan Teng, M.S.
LeighAnn Kowalski, M.D.
Daine Stevens, M.S.
Adam Young, M.S.
Ryan Tubman, M.S.
Kevin Oelstrom, Ph.D.
Tim Requarth, Ph.D.
Anup Patel, M.D.
Jeff Guzelian, M.D.
Natalie Boone, M.S.
2020-2022
2020-2022
2019-2021
2019-2020
2019
2017-2018
2017-2018
2016-2017
2015-2016
2015-2016
2014-2016
2012-2014
2011-2012
2011
2008-2011
2010
2009
2009
2007-2008
2006-2007
2006-2007
2002-2003
Investment Associate, Hi2 Global, New York City, NY
Ph.D. Student, Biological Studies, Vanderbilt University, Nashville, TN
Ph.D. Candidate, Interdepartmental Neuroscience Program, Northwestern University, Evanston, IL
Ph.D. Graduate Student, University of California, Irvine, CA
Law Clerk, SKH Family Law and Estate Planning, Portland, OR
Ph.D. Grad Student, Interdepartmental Neuroscience Program, Northwestern University, Evanston, IL
Resident, Neurology, University of Illinois College of Medicine, Chicago, IL
Biologics MSAT Engineer, AGC Biologics, Hillsboro, OR
Investigator, Genome Editing, GSK, Wayne, PA
Freelance Software Engineer, San Francisco, CA
Freelance Tutor, Houston, TX
Assistant Professor, General Medicine, Rush University, Chicago, IL
Project Manager, WuXi AppTec, Shanghai, China
Physician at OSF HealthCare St. Elizabeth Hospital and Medical Group, IL
Teacher, Josef-Schwartz-Schule, Greater Heilbronn Region
Laboratory Scientist, Cellular Therapy, Versiti Blood Center of Michigan, Grand Rapids, MI
Partner, ClearView Healthcare Partners, San Francisco, CA
Product Manager, Cell Health Analysis, Promega Corporation, Madison, WI
Research Professor, Neuroscience & Physiology, NYU Grossman School of Medicine, New York City, NY
Resident, Anesthesiology, Northwestern Memorial Hospital, Chicago, IL
Diagnostic Radiology Specialist, Florida Cancer Specialists and Research Institute, Tampa, FL
Senior Director Medical Writing, Oncology, Astellas Pharma, Chicago, IL
Selected Publications
Join the Lab
Contact us
width=”100%” height=”600″ frameborder=”0″ allowfullscreen=”allowfullscreen” aria-hidden=”false”>